Many physicians turn to the FDA-approved drug label (package insert) for information about genetic testing that can be used to optimize drug response, also known as pharmacogenomics/pharmacogenetics (PGx). Information about PGx appears in the labels of hundreds of drugs, but how reliable and useful is this information?
This question has taken on new importance recently as the FDA has stepped in to more actively regulate PGx tests. Many test providers have responded by removing information about specific drugs from their test reports and only reporting genotypes, leaving the drug label as the primary source for PGx-related prescribing information.
According to a safety communication issued by the FDA in Oct 2018, PGx testing labs should limit their PGx offerings to those drugs that have FDA-authorized labels describing how genetics can be used to inform treatment. Moreover, the FDA strongly recommends that clinicians consult the FDA-approved drug label for PGx information in the course of patient care.
Given the emphasis on the drug label for both regulation and clinical use of PGx, it deserves some scrutiny. In this post, I review the history of PGx information in the drug label, explore the validity and utility of that information and raise concerns about relying on PGx information in the FDA drug label to make treatment decisions.
A brief history of PGx in the FDA drug label
From early on in the genomics era, the FDA seemed to champion the use of genetic information to guide use of therapeutics. A guidance for industry published in 2005 encouraged voluntary submission of PGx data during the drug development process.
The 2005 FDA guidance provided the first systematic framework for developing and approving PGx information and has resulted in voluntary submission of PGx data to the FDA and co-development of pharmacogenomic tests for many drugs, especially in the area of oncology.
After a drug is on the market, re-labeling to add PGx information can be initiated by the FDA, academic researchers or the pharmaceutical developer, but there is a high threshold for revisions. The motivation for re-labeling is rooted in supporting the safety of the drug, as outlined in FDA regulation CFR 201.57. This regulation states that the drug label should describe the evidence and identify the test necessary to select patients that are at a safety risk.
Post-approval re-labeling practices were promoted in the mid 2000s to encourage addition of PGx data relevant to safety and efficacy. During this time, over a dozen labels were revised, but these efforts have significantly tapered off in recent years. Moreover, addition of PGx information to the label in the post-approval setting can be a lengthy process, ranging from 15-44 years after drug approval, unless there’s a serious safety issue (Vivot et al 2015). As a result, clinically valid PGx tests for some drugs are not mentioned in the drug label.
A survey of drugs with PGx in the FDA label
Although some PGx information has appeared in the labeling of drugs approved prior to 1995, most PGx biomarker labeling occurred after 1996. (Freuh et al. 2008).
PGx biomarkers can include a patient’s germline genotype, the genetics of a pathogen (specifically for infectious disease) or somatic variation in a tumor (in the case of cancer).
Over 260 drug-gene pairs are found in drug labels, a large proportion of which are for cancer therapies. For many of these, somatic alterations in the tumor are biomarkers of drug efficacy and testing is often required prior to prescribing.
PGx tests based on germline (inherited) variation are found across a wide range of therapeutic areas. These tests often involve examining genetic variation in drug metabolism, including the CYP450 genes, which in turn influences the drug’s efficacy or side effects.
PGx information contained in the FDA drug label
Because physicians rely on the FDA-approved drug label for PGx information (Stanek et al 2013), it’s important to take a critical look at what gets included in that label. According to the 2005 FDA guidance, PGx data may be included in the drug label for any one of a number of reasons:
· To choose a dose
· To identify patients at risk
· To identify responders
· Simply for informational purposes
The purpose of including PGx information in the drug label is either to alert the physician that genetic testing is ‘required’ or ‘recommended’, or simply for ‘informational’ purposes. The FDA, in its Table of Pharmacogenomic Biomarkers in Drug Labeling, used to categorize each drug’s PGx information in this way (required, recommended, informational), but in 2009 that system was abandoned. However, a similar annotation was later adopted by the PGx knowledgebase, PharmGKB.
The vast majority of drugs with PGx information in the label include PGx for informational purposes only. Less than 15% of labels for drugs with germline PGx information suggest that testing is ‘recommended’ or ‘required’. That number is higher for oncology drugs that use somatic testing of the tumor, where ~60% are recommended or required.
For those drugs where PGx testing is either required or recommended, the language is clear and direct, describing what testing should be done and how to alter treatment based on the test results:
‘All patients should be screened for the HLA-B*5701 allele prior to initiating therapy with ZIAGEN or reinitiation of therapy with ZIAGEN, unless patients have a previously documented HLA-B*5701 allele assessment. ZIAGEN is contraindicated in patients: who have the HLA-B*5701 allele.’
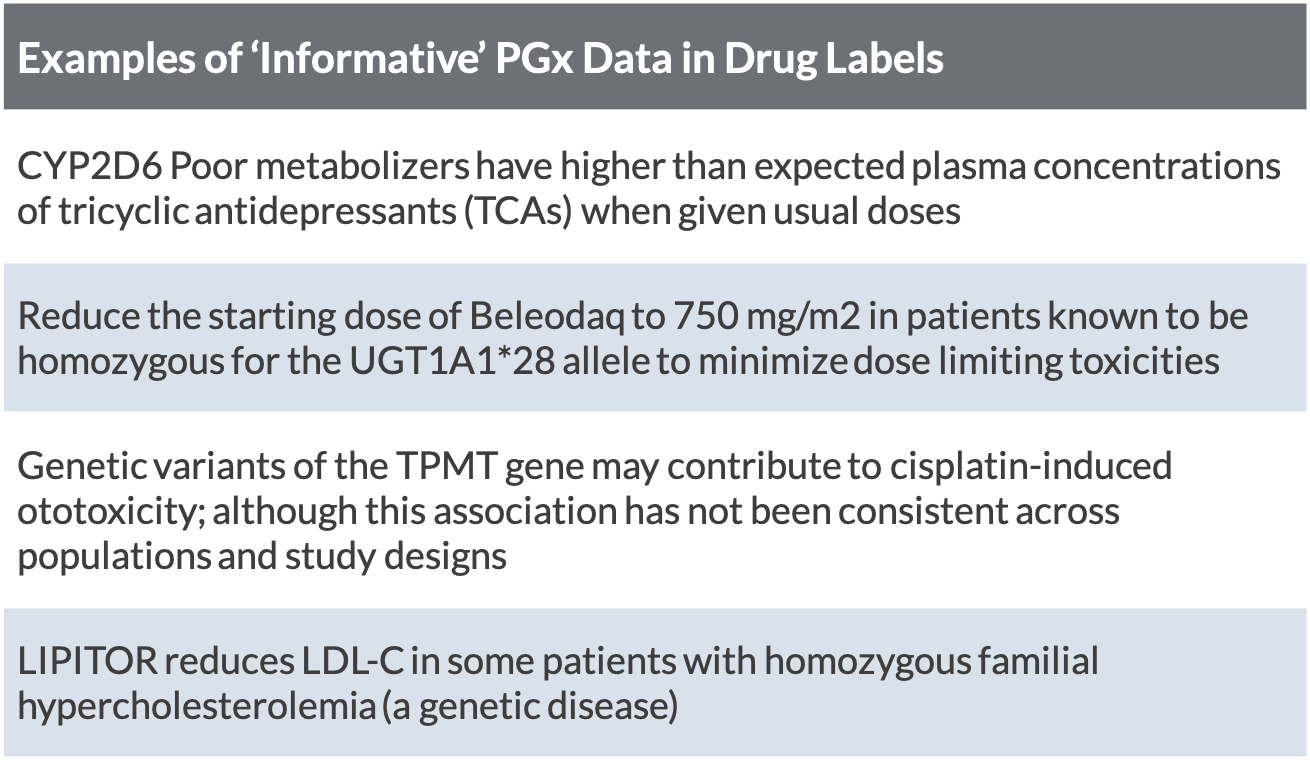
The interpretation of PGx information found in drug labels for ‘informational’ purposes is less straightforward. These labels can include PGx information for a number of reasons, including to: define the disease; describe the drug’s mechanism of action, drug interaction, change in pharmacological endpoints, increased toxicity or decreased efficacy; or simply inform that there is no change related to the gene (Vivot et al 2015).
For some of these drugs, the evidence supporting the association between the PGx biomarker and treatment response is not convincing (e.g. third row in table). For others, the evidence may be convincing, but limited availability of PGx tests at the time of re-labeling made it difficult for the FDA to be prescriptive about requiring or recommending testing (Drozda 2018).
The result is that physicians who read these labels are left not knowing whether to test their patients for these pharmacogenes or not, and if they do test, how the test results should inform use of the drug.
Clinical validity and utility of PGx in drug labels
Genetic tests are generally evaluated based on their analytical validity, clinical validity and clinical utility. As described in a previous blog post, clinical validity includes scientific evidence backing associations between genetic variants and clinical outcomes, while clinical utility includes evidence that the test results change medical practice.
According to the 2005 FDA guidance, a PGx biomarker included for informational purposes in the drug label would need to be a known, valid biomarker, defined by having: 1. analytical validity (assay accurately detects what it says it detects); 2. evidence of genetic association between gene and drug-related phenotype (e.g. clinical validity); and 3. accepted in the broad scientific community.
This FDA guidance suggests that PGx biomarkers in the drug labels should be clinically valid, and yet according to two research studies, less than half of drug labels with pharmacogenomic information provide convincing evidence of clinical validity (Chin et al. 2017; Wang et al. 2014). This incongruity may be explained by evaluators using different data, or different metrics for establishing clinical validity.
Frameworks for establishing clinical validity of genetic tests like the EGAPP framework from CDC, used as a benchmark in the aforementioned research studies, have been around since 2005. These frameworks consider the body of epidemiological evidence relating the genotype with phenotype, which is generally a clinical outcome.
Some would argue that, especially for the CYP450 drug metabolizing enzymes, evidence linking P450 genotype to a clinical outcome may not be necessary. Instead, knowing a person’s metabolizer status and how that should impact the level of the drug is enough to extrapolate the need for dosing adjustment.
It’s not clear what, if any formal framework is used by the FDA to evaluate the clinical validity of gene-drug response associations for inclusion in the drug label.
Furthermore, data used for making assertions of clinical validity may come from the published literature, which is found in the public domain, or private data submitted by the test manufacturer at the time of drug approval. If private data are not included in the label, it’s difficult to judge the clinical validity.
Moreover, as data (evidence) accumulates over time, assertions of clinical validity may change and yet there is no way to capture or convey these changes in a timely fashion, although there have been instances where PGx information used to be in a drug label but was later removed.
Dangers of relying on FDA drug labels for PGx
There are several inherent dangers to relying solely on information contained in the FDA-approved drug label for making PGx-related treatment decisions.
Drug labels missing PGx information
First, some valid PGx biomarkers of drug response do not appear in the FDA label. For example, the drug allopurinol (Zyloprim®) was approved in 1966 to treat gout and carries of risk of life-threatening severe cutaneous adverse reaction (SCAR), especially in people with a variant of HLA-B found most commonly in Asian populations. CPIC guidelines published in 2016 provide evidence of clinical validity of the PGx association between HLA-B*5801 and allopurinol-induced SCAR. The drug label was most recently updated in Jan 2019, but still does not contain information about the increased risk of SCAR in patients with the HLA-B*5801 genotype.
In a previous blog post, I noted that among the most clinically valid 35 gene-drug pairs (according to PharmGKB and CPIC), eight (23%) are not yet mentioned in their respective drug labels.
Drug label with PGx information misaligned with available evidence
Second, some PGx markers in the drug label recommend changes to prescribing but these recommendations are not supported by available scientific evidence.
One example is the drug label for the Hungtington’s disease drug tetrabenazine and the gene CYP2D6. The label states that patients should be genotyped for CYP2D6, and if they are poor metabolizers, they are advised to use a lower dose of the drug. There is only one published study in the literature, and it failed to find an association between CYP2D6 poor metabolism and response to the drug (Raja et al. 2013).
The evidence used by the FDA to make the recommended dosing change is not provided in the drug label. The label only states that the pharmacokinetics of the drug in CPY2D6 poor metabolizers has ‘not been systematically evaluated,’ but that it’s likely that the active metabolites of the drug would be increased, similar to as if the patient had taken a strong CYP2D6 inhibitor. In other words, there doesn’t appear to be any clinical evidence of association between CYP2D6 poor metabolism and adverse outcomes, or even a pharmacokinetic analysis of CYP2D6 poor metabolism.
Perhaps in this case, the risk of adverse events due to overexposure of the drug’s active metabolites is significantly greater than the risk of under-dosing to warrant making a dosing adjustment based on purely mechanistic rationale. Following this logic, all drugs metabolized by polymorphic P450 genes might qualify for dosing adjustments based simply on metabolizer status, and yet we don’t find PGx information in the label of all of those drugs.
This example highlights a lack of transparency in the supporting evidence (or lack of evidence) used to make PGx assertions and suggests a lose framework at best for evaluating clinical validity of PGx biomarkers.
Recommendations
The current PGx ecosystem includes professional organizations like CPIC and PharmGKB playing important roles as evidence curators and interpreters of PGx information in the drug label. Clinical laboratories offering testing have found it helpful to customers to include prescribing information in their reports. This is as much a matter of convenience to the healthcare provider as a necessity for conveying clear prescribing guidance that physicians may not get from the drug label.
Regulators, in an attempt to reign in this system, may have inadvertently made PGx tests less safe as testing labs scramble to conform to their demands with stripped down versions of their test reports.
In the current climate, it may be time to re-evaluate the drug label and re-establish its central role as a principal, trusted source of prescribing information related to PGx. This could be achieved by developing new standards for inclusion of PGx information in the label and a standard language for conveying prescribing changes.
If PGx biomarkers must be clinically valid to be included in the label, a framework for establishing clinical validity needs to be articulated and applied evenly. As the field of genetic testing has matured, such frameworks are becoming more commonplace and provide much needed structure to evidence evaluation.
Much of the discord between the FDA and PGx test developers is related to a lack of consensus of what constitutes a clinically valid PGx test and what information testing labs should convey to patients. A new guidance for PGx information in the FDA drug label could prove to be the key to safer and more effective therapies through PGx testing.
Learn More
Precision Medicine Advisors specializes in communicating precision medicine to lay professional audiences, providing scientifically sound, unbiased information to promote the responsible use of genomics in medicine.
To improve your genomic literacy, check out our online courses at precisionmedicineacademy.org.
Contact info: jeanette@pmedadvisors.com